Paul Salvaterra, Ph.D.

Duarte Cancer Center
Duarte, CA 91010
Ph.D., Biochemistry State University of New York, Buffalo, 1973
Alexander Von Humboldt Foundation
Department of Psychiatry, Martinsried, Munich, Federal Republic of Germany, 1985Chemistry Department, Indiana University, Bloomington
1974, 1976-1977
Neuronal Gene Expression
We are interested in genes that determine the neuronal cellular fate and specific neurotransmitter phenotypes of cells in the nervous system. The reasons for our interest are two fold. First, identification of the genetic pathways operating in specific types of neurons could explain why they die in neurodegenerative diseases such as Alzheimer’s, Parkinson’s and amyotrophic lateral sclerosis. All of these disorders have in common the property that only certain types of neurotransmitter specific neurons degenerate. Secondly, neuronal cell replacement therapies using differentiated stem cell progeny hold the potential to reverse the devastating consequences of neurodegenerative diseases. The genetic personality of specific kinds of neurons must first be defined in order to use proper cells for neuronal replacement and this information will be essential in directing stem cell differentiation into proper developmental pathways.
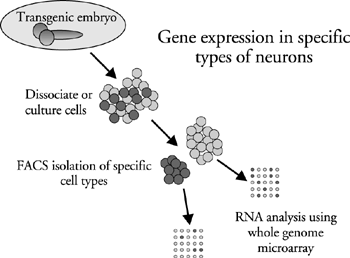
Our current approach to the problems of specification and neurotransmitter phenotype is to create transgenic animals where different neurotransmitter phenotypes are labeled with a fluorescent reporter gene. Labeled neurons are then isolated using Fluorescence Activated Cell Sorting. The purified populations of neurons are analyzed for their whole genome expression patterns using DNA microarray technology. We have successfully applied this approach using Drosophila cholinergic neurons and are now extending our observations to other classes of neurons that use GABA or glutamate as neurotransmitters. Cholinergic neurons express unique sets of ion channels, receptors and other types of genes. We also see unique sets of transcriptional regulatory proteins, and these may be important in the developmental pathways that result in the production of cholinergic neurons. The transcriptional regulatory properties of the cholinergic gene locus is an additional focus of the lab and our approach is mainly molecular biological using transgenic animals where regulatory factors and cis regulatory DNA elements are identified and tested.
A second project in the laboratory involves DNA microarray analysis of animals carrying mutations in the Presenilin gene. The normal function of Presenilin is unknown but mutations in this gene are the major cause of familial Alzheimer’s disease in humans. Using Drosophila as a model genetic organism, we have identified lysosomal dysfunction as one major consequence of animals having Presenilin loss-of-function alleles. Biochemical analysis has confirmed some of the gene expression results and we hope to expand this study to transgenic mice in the near future. Our results will not only help define genetic pathways compromised in Alzheimer’s disease but could also help identify function(s) for the Presenilin protein.
Strategy used to determine the complete genetic profile of neurons using different neurotransmitter phenotypes
Transgenic lines with fluorescent reporter genes are constructed using cis DNA regulatory elements from neurotransmitter specific genes. Neurons are then obtained from either dissociated nervous system or they are generated by culturing neural progenitor cells. The neurons are purified into neurotransmitter specific populations on the basis of their fluorescence. In Drosophila we then extract RNA from the purified neurons and determine expression using commercially available whole genome microarrays.