
Zhaohui Gu Lab
Research Highlights
Precision bioinformatics software development
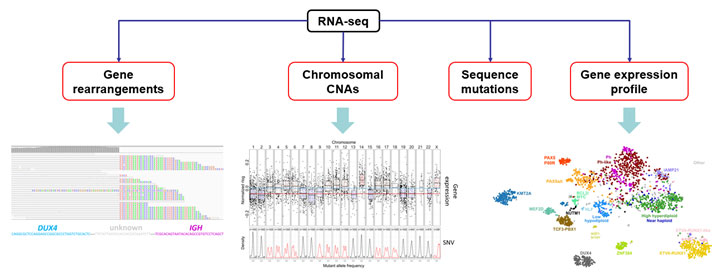
Through analyzing the largest leukemia genomics dataset, Dr. Gu developed a robust RNA-seq based ALL classification system, which employs both the driver genetic lesions and the signature gene expression profile to reliably assign leukemia subtypes. However, the implementation of the system for public use is still challenging due to the lack of disease-specific analysis tools. To bridge the gap in customized genomic data analysis, the Gu Lab will focus on developing bioinformatics tools with specific optimization and integration of prior disease knowledge to provide optimal results for blood cancer studies.
Functional genomics and epigenomics study of novel leukemia subtypes
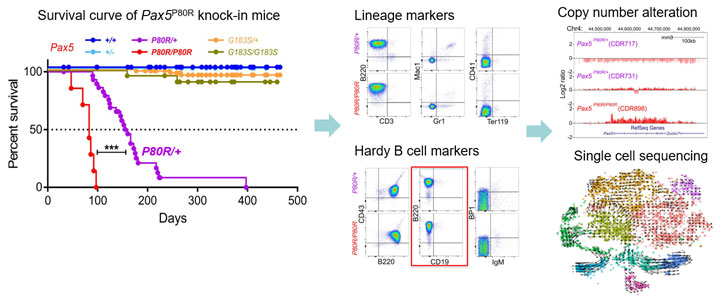
PAX5 is a key transcription factor in B-cell development and commitment. PAX5 P80R is an exemplar of a single point mutation as an initiating driver in ALL, but its role in leukemogenesis compared to other PAX5 mutations on the same DNA binding domain is largely unknown. Through mouse model, we have shown that Pax5 P80R can initiate B-lineage leukemia through the development of other somatic alterations, such as deletion of the wild-type allele of Pax5 in Pax5P80R/+ mice, which indicates a mutagenesis evolution was involved in this specific leukemia subtype. The Gu lab will apply multiple functional genomics and epigenomics platforms, such as ChIP-seq, ATAC-seq, CUT&RUN/CUT&Tag etc. to study the unique role of PAX5 P80R versus the other PAX5 sequencing mutations, and elucidate the step-wise mutagenesis and immune response evolution during leukemogenesis using single-celling (sc-) analysis platforms (scRNA-seq, sc-RNA-seq with genotyping, CyTOF etc.)
An Assistant Professor in the Department of Computational and Quantitative Medicine and the Department of Systems Biology, Zhaohui Gu, Ph.D. conducts research that focuses on identifying novel leukemia subtypes and the driver genetic lesions, developing clinically applicable leukemia classification system, developing machine-learning-based analysis tools for leukemia/cancer genomics studies, studying the mechanism of leukemogenesis in each disease subtype and developing therapeutic strategies for each leukemia subtype.



Helen Han, M.S., graduated from UCLA with a bachelor’s degree in 2016 and from Charles R. Drew University of Medicine and Science with a master’s degree in 2018. In September 2020, she joined the Zhaohui Gu laboratory in the Department of Computational and Quantitative Medicine and Department of Systems Biology.
Currently, Han works with Zhilian Jia, Ph.D., to understand the role and function of novel Pax5 mutant-driven subtypes of ALL in regulating leukemogenesis. Outside of the lab, she loves to go hiking and travel whenever time permits.
Degrees
- 2018, M.S. Biomedical Sciences, Charles R. Drew University of Medicine and Science, Los Angeles, CA
- 2016, B.S. Molecular, Cell and Development Biology, UCLA, Los Angeles, CA
Professional Experience
- 2020-present, Research Associate II, Department of Computational and Quantitative Medicine, City of Hope, Monrovia, CA
- 2019-2020, Research Associate I, Department of Systems Biology, City of Hope, Monrovia, CA
Awards
- 2014, American Red Cross Blood Drive Scholarship


Zunsong Hu, Ph.D., received his bachelor’s degree in preventive medicine in 2012 from Huazhong University of Science and Technology in Wuhan, China. Then, he joined Dr. Dongfeng Gu’s laboratory in Fuwai Hospital, Peking Union Medical College in Beijing, China, for his Ph.D. training in epidemiology and health statistics and graduated in 2017.
He joined the laboratory of Dr. Qi Zhao, M.D., Ph.D., in the University of Tennessee Health Science Center for his postdoctoral training in 2018, and then he joined the laboratory of Zhaohui Gu, Ph.D., in the Department of Computational and Quantitative Medicine and Department of Systems Biology in September 2020.
His major scientific interests are integrating multi-omics datasets to study complex diseases and risk factors. Currently, he is using high-throughput omics (including genomics, bulk and single-cell transcriptomics and functional genomics) to develop classification standards for leukemia and cancer subtypes. His research aims are to dissect genomic, transcriptomic and epigenomic mechanisms underlying each malignancy subtype.
Degrees
- 2017, Ph.D., Epidemiology and Health Statistics, Peking Union Medical College, Beijing, China
- 2012, B.S., Preventive Medicine, Huazhong University of Science and Technology, Wuhan, China
Professional Experience
- 2020-present, Postdoctoral Fellow, Beckman Research Institute of City of Hope
- 2017-2020, Postdoctoral Research Associate, University of Tennessee Health Science Center
Awards
- 2019, Member, American Society of Human Genetics
- 2019, Member, American Heart Association
Publications
- Hu Z, Tylavsky FA, Kocak M, Fowke JH, Han JC, Davis RL, LeWinn KZ, Bush NR, Sathyanarayana S, Karr CJ, et al. Effects of Maternal Dietary Patterns during Pregnancy on Early Childhood Growth Trajectories and Obesity Risk: The CANDLE Study. Nutrients 2020;12.
- Hu Z, Tylavsky FA, Han JC, Kocak M, Fowke JH, Davis RL, Lewinn K, Bush NR, Zhao Q. Maternal metabolic factors during pregnancy predict early childhood growth trajectories and obesity risk: the CANDLE Study. International Journal of Obesity 2019; 43:1914-1922.
- Hu Z, Liu F, Li M, He J, Huang J, Rao DC, Hixson JE, Gu C, Kelly TN, Chen S, Gu D, Yang X. Associations of variants in the CACNA1A and CACNA1C genes with longitudinal blood pressure changes and hypertension incidence: The GenSalt Study, Am J Hypertens, 2016, 29(11): 1301-1306.
- Han C, Hu Z, Liu F, Yang X, Kelly TN, Chen J, Huang J, Chen CS, He J, Chen S, Wu X, Gu D, Lu X. Genetic variants of cGMP-dependent protein kinase genes and salt sensitivity of blood pressure: the GenSalt study, J Hum Hypertens, 2018; 2018;33:62-68.
- Li L, Wang L, Li H, Han X, Chen S, Yang B, Hu Z, Zhu H, Cai C, Chen J, Li X, Huang J, Gu D. Characterization of LncRNA expression profile and identification of novel LncRNA biomarkers to diagnose coronary artery disease. Atherosclerosis, 2018, 275:359-367.
- Li H, Han X, Hu Z, Huang J, Chen J, Hixson JE, Rao DC, He J, Gu D, Chen S. Associations of NADPH oxidase-related genes with blood pressure changes and incident hypertension: The GenSalt Study, J Hum Hypertens, 2018, 32(4): 287-293.
- Han X, Hu Z, Chen J, Huang J, Huang C, Liu F, Gu C, Yang X, Hixson JE, Lu X, Wang L, Liu D-P, He J, Chen S, Gu D. Associations between genetic variants of NADPH oxidase-related genes and blood pressure responses to dietary sodium intervention: The GenSalt Study, Am J Hypertens, 2017, 30(4): 427-434.
- Cheng M, Hu Z, Lu X, Huang J, Gu D. Caffeine intake and atrial fibrillation incidence: dose response meta-analysis of prospective cohort studies, Can J Cardiol, 2014, 30(4): 448-54.


Zhilian Jia, Ph.D., received her bachelor’s degree in nursing in 2009 from Peking University Health Science Center in Beijing, China. Then she joined Dr. Qiang Wu’s lab for her Ph.D. training in Shanghai Center for Systems Biomedicine, Shanghai Jiao Tong University in Shaghai, China.
She received her Ph.D. in molecular biology in 2015 and began her postdoc training in pursuing the regulatory mechanism of the architectural protein CTCF.
She joined the lab of Zhaohui Gu, Ph.D., in 2021 to study the functional impact of the driver genetic lesions in ALL. Her current projects are using in vitro and in vivo models, coupled with multiple functional genomics platforms, to study the role of different Pax5 mutations in driving leukemogenesis.
Degrees
- 2015, Ph.D., Molecular Biology, Shanghai Jiao Tong University, Shanghai, China
- 2009, Nursing, Peking University, Beijing, China
Professional Experience
- 2021-present, Postdoctoral Fellow, Beckman Research Institute of City of Hope, Duarte, CA
- 2017-2020, Research Fellow, Shanghai Jiao Tong University, Shaghai, China
- 2015-2017, Postdoctoral Fellow, Shanghai Jiao Tong University, Shanghai, China
Awards
- 2018, First prize of Natural Science Award of Ministry of Education of China
- 2018, First prize of Natural Science Award of Shanghai
- 2015, Outstanding Graduates of Shanghai City
- 2015, National Scholarship for Excellent Students
- 2013, Fung Scholarship
- 2011, Koguan Encouragement Scholarship
- 2007, Outstanding Student Award for Medicine
Publications
- Jia Z*, Li J*, Ge X*, Wu Y, Guo Y, Wu Q. Tandem CTCF sites function as insulators to balance spatial chromatin contacts and topological enhancer-promoter selection. Genome Biology. 2020; 21. Doi: 10.1186/s13059-020-01984-7.
- Jia Z, Wu Q. Clustered Protocadherins Emerge as Novel Susceptibility Loci for Mental Disorders. Front Neurosci. 2020 Nov 12;14:587819.
- Wang N, Jia Z, Wu Q. RFX5 regulates gene expression of the Pcdha cluster. Hereditas (Beijing). 2020 Aug 20;42(8):760-774.
- Wu Q, Jia Z. Wiring the Brain by Clustered Protocadherin Neural Codes. Neuroscience Bulletin. 2020 Sep 17. doi: 10.1007/s12264-020-00578-4
Wu Y, Jia Z, Ge X, Wu Q. Three-dimensional genome architectural CCCTC-binding factor makes choice in duplicated enhancers at Pcdhα locus. Science China Life Science. 2020; 63. Doi: 10.1007/s11427-019-1598-4. - Lu Y*, Shou J*, Jia Z, Wu YH, Li JH, Guo Y, Wu Q. Genetic evidence for asymmetric blocking of higher-order chromatin structure by CTCF/cohesin. Protein Cell. 2019; 10: 914-920.
- Shen X, Lu Y, Jia Z, Wu Q. N-WASP regulates cortical neuron migration through its polyPro and VCA domains. Hereditas (Beijing). 2018; 40(5): 390-401.
- Shu L, Jia Z, Wu Y, Suo L, Jia L. The application of CRISPR/Cas9 in brain research. Acta Veterinaria Zoolechnica Sinica. 2016; 47(7): 1316-1323
- Guo Y*, Xu Q*, Canzio D, Shou J, Li J, Gorkin D, Jung I, Wu H, Zhai Y, Tang Y, Lu Y, Wu Y, Jia Z, Li W, Zhang MQ, Ren B, Krainer AR, Maniatis T, Wu Q. CRISPR inversion of CTCF sites alters genome topology and enhancer/promoter function. Cell. 2015;162(4): 900-910.
- Li J*, Shou J*, Guo Y, Tang Y, Wu Y, Jia Z, Zhai Y, Chen Z, Xu Q, Wu Q. Efficient inversions and duplications of mammalian regulatory DNA elements and gene clusters by CRISPR/Cas9. Journal of Molecular Cell Biology. 2015: p. mjv016.
- Jia Z, Guo Y, Tang Y, Xu Q, Li B, Wu Q. Regulation of the protocadherin Celsr3 gene and its role in globus pallidus development and connectivity. Molecular and Cellular Biology. 2014; 34: 3895-391
We collaborate with organizations in progressing the development of new treatments in our specialized areas of research.

Latest Research News
No articles found matching the selected criteria.
34.1293487, -117.9726643
Duarte , CA 91010