Ren-Jang Lin, Ph.D.

Duarte Cancer Center
Duarte, CA 91010
1983, Ph.D., Biological Chemistry, Pennsylvania State University
1978, B.S., Agricultural Chemistry, National Taiwan University
1983‐1986, Biology, California Institute of Technology (with support from American Cancer Society and National Institutes of Health), Pasadena, CA
2012–present, Associate Dean for Curriculum Development, Irell & Manella Graduate School of Biological Sciences at City of Hope, Duarte, CA
2000–present, Professor, Center for RNA Biology and Therapeutics, Beckman Research Institute, City of Hope, Duarte, CA
2016–present, Distinguish Guest Professor, Fujian Medical University Union Hospital, Fuzhou, China
1998‐2012, Adjunct Research Professor of Biochemistry and Microbiology, Loma Linda University, Loma Linda, CA
1995‐2000, Associate Professor of Molecular Biology, Beckman Research Institute, City of Hope, Duarte, CA
1993‐1995, Director, Department of Molecular Biochemistry, Beckman Research Institute, City of Hope Duarte, CA
1987‐1993, Assistant Professor of Microbiology, University of Texas, Austin, Texas
Overview:
Non-coding intervening sequences, or introns, interrupt most eukaryotic genes. Cells must remove introns with great precision from these gene transcripts in order to accurately express the genetic information. Intron removal is done through a process called RNA splicing, and the spliceosome is a ribonucleoprotein complex that splices pre-mRNAs through two transesterification reactions. In addition to being highly precise, the spliceosome is also quite flexible; it can splice different sets of expressing sequences, or exons, together to produce multiple splice variants from one gene transcript. Alternative splicing allows human cells to produce a million of proteins from their 50,000 genes. Thus, an abnormal or defective spliceosome would bring disease or even death to an organism. Our goals are to understand the molecular detail of the spliceosome in model organisms as well as to study its function and regulation in normal and cancerous cells.
Human DHX16/PRP2 protein in splicing and cell death
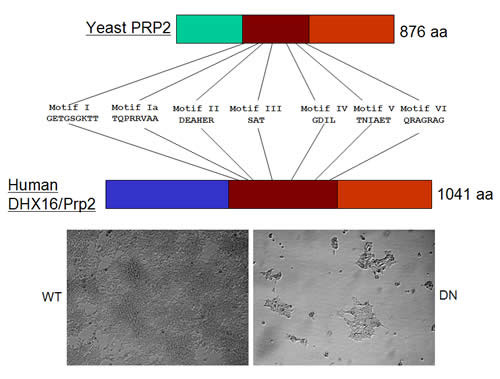
Spliceosome requires different DExD/H-box RNA helicases at assembly, activation, and disassembly steps. Mutations in these helicase genes of S. cerevisiae could abolish splicing and cell growth; however, their effect in humans is less known. S. cerevisiae Prp2 protein is a late spliceosomal helicase; prp2 mutants accumulate unspliced pre-mRNA in vivo and their extracts form an inactive pre-catalytic spliceosome in vitro (see project 2). We showed that an siRNA-mediated knockdown of human DHX16 gene expression caused a quick and drastic decrease of mRNAs from several intron-containing genes. Overproduction of the protein mutated at its helicase domain had a similar effect. In both cases, the unspliced pre-mRNA from a reporter was detected. Splicing in HeLa extracts was inhibited by an antibody against a Dhx16 peptide; however, spliceosomes were nonetheless formed. Thus, sequence and function similarities indicate that human Dhx16 is likely the orthologue of S. cerevisiae Prp2. Interestingly, the antibody immunoprecipitated from splicing reactions the spliceosomal snRNAs, unspliced pre-mRNA, and the splicing intermediates. Yeast two-hybrid screens revealed an interaction between hPrp2 and a second-step splicing factor further suggesting a potential role of hPrp2 in spliceosome transition. We have established a stable cell line having a controllable dominant negative hPRP2; upon induction, DNA fragmentation occurred and cells died of apoptosis. This provides an opportunity to examine how splicing, transcription, and cell growth respond to a stall of spliceosome in vivo.
Finding alternative splicing variants in breast cancer
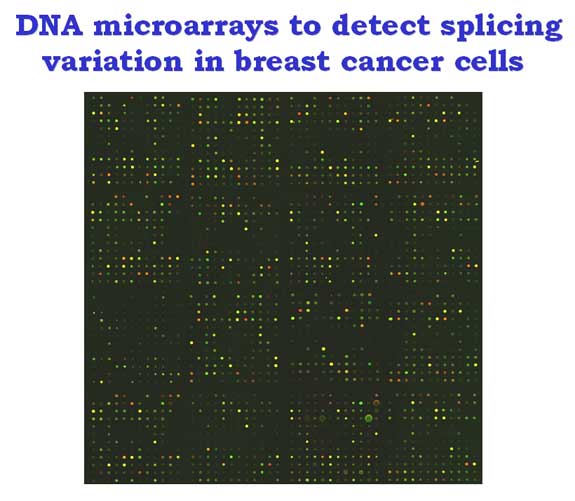
Growing evidence indicates that alternative or aberrant pre-mRNA splicing takes place during the development, progression, and metastasis of breast cancer. However, what splicing changes are really important in tumorigenesis and cancer biology remains to be elucidated.
We used microarrays of junction oligonucleotides representing 120 alternative exons of 64 genes to detect splicing differences between breast cancer cells and normal human mammary epithelial cells. Several splicing alterations in genes including CD44, FAS, APLP2 and MYL6 were detected by microarrays and RT-PCR.
We also compared two breast cancer cell lines MCF7 (estrogen receptor positive) and MDA-MB-231 (estrogen receptor negative) cultured in two-dimensional (2D) flat dish or in three-dimensional (3D) Matrigel. Interestingly, only a subset of the splicing alterations between MCF7 and MDA-MB-231 cells detected in 2D samples was retained in 3D samples, suggesting culturing conditions affect splicing variations.
We further compared splicing in MCF7 cells grown in dish, Matrigel, or xenograft in mouse and found at least one example that splicing variants expressed in xenograft could be identified in 3D but not in 2D cultures. Thus, our preliminary investigation indicates that microarrays can be used to identify splicing variants that potentially can be used as breast cancer markers.
The catalytic core of the spliceosome
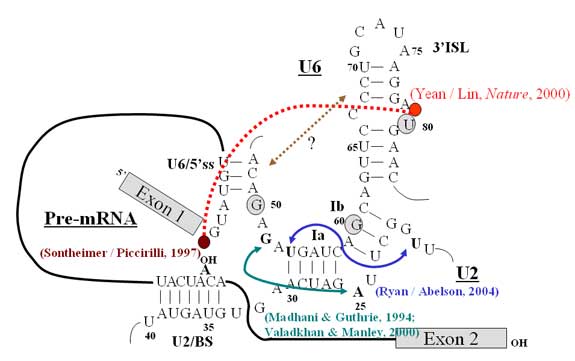
The spliceosome is composed of five small nuclear RNAs and about a hundred of proteins, whose function is to recognize and splice exons together. A longstanding question about spliceosome function is whether snRNAs or proteins catalyze the splicing reaction. Since the spliceosome is a metalloenzyme, finding the spliceosomal components that coordinate catalytic metals is crucial in addressing the question. There is evidence of genetic and biochemical nature indicating that the U6 snRNA is at the catalytic core of the spliceosome.
By reconstituting the yeast spliceosome with a phosphorothioate-substituted U6 snRNA, we demonstrated that the 5’ phosphate of the eightieth nucleotide (U80) of U6 coordinates a catalytically important magnesium ion for the first step of splicing. We found, using similar approaches, that phosphates of A59 and G60 of U6 snRNA are also critical for the catalytic activity of the spliceosome. Thus, these observations support the idea that U6 snRNA is not only at the core of the spliceosome but also directly participates in the chemical reaction of splicing, and that the spliceosome may have an evolutionary root in RNA-based machinery.
Yeast splicing extracts preparation
Extract preparation for in vitro pre-mRNA splicing. YPD cultures of S. cerevisiae mutant strain 3.2 A1D [MATa, prp2-1, ade2, his3, lys2-801, ura, first described in 19991] were grown at 26°C until they reached late logarithmic phase (A600 of 3-5). 2 mls of the protease inhibitor phenylmethyl sulfonyl fluoride [0.5M PMSF] per liter of culture were added and the culture was allowed to grow for another 30 min.
Cells were collected by centrifugation at room temperature for 10 min at 7000 (3800 x g) in a Sorvall GS3 rotor. The wet weight of the cells (30g) were measured and this value was used for the rest of the extract preparation procedure. Pellets were re-suspended in 150mls of TE Buffer [50mM Tris-HCl (pH 8.0), 0.2mM EDTA (pH 8.0)] at 50mls per 10g of cells.
Cells were collected by centrifugation at room temperature for 6 min at 6000 (2800 x g) in a GS3 rotor. Pellets were then re-suspended in 90mls of SB30 buffer [1M Sorbitol, 50mM Tris-HCl (pH 7.8), 10mM MgCl2, 30mM DTT] at 30 mls per 10g of cells. Cells were collected by centrifugation at room temperature for 6 min at 6000 rpm (3900 x g) in a Sorvall GSA rotor. Pellets were next re-suspended in 60mls of SB3 buffer [1M Sorbitol, 50mM Tris-HCl (pH 7.8), 10mM MgCl2, 3mM DTT] at 20mls per 10g of cells. 10ul of suspended cells were added to 1ml of 10% SDS and the A800 was measured.
The suspended cells were treated with the supernatant from the addition of 9 mg of zymolyase (from ICN, TM-100T)in 600ul of zymolyase buffer [20mM KPO4 (pH 7.8), 5% glucose] at 3 mg and 200ul per 10g of cells, respectively. The cells were shaken at 30°C for 75 min at 120 rpm and then the A800 was re-measured by adding 10ul of the zymolyase-treated cell suspension plus 1ml of 10% SDS. Before proceeding, the A800 of the suspension must decrease by >80% from the initial absorbance reading.
Zymolyase (from Seikagaku) treated cells, called spheroblasts, were collected by centrifugation at room temperature for 5 min at 3000 rpm in a GSA rotor. The pellets were gently re-suspended in 90mls of SB3 containing 0.5mM PMSF at 30mls per 10g of cells. The spheroblasts were collected at room temperature for 5 min at 3000 rpm in a GSA rotor. Spheroblast pellets were chilled on ice for 30 min.\
Pellets were re-suspended in 30mls of buffer A containing protease inhibitors [10mM HEPES (pH 7.8), 1.5mM MgCl2, 10mM KCl, 0.5mM DTT, 1uM leupeptin, 1uM pepstatin, 1um benzamidine, 1uM PMSF] at 10mls per 10g of cells. Re-suspended pellets were poured into a dounce glass homogenizer (type B) and lysed by 14 hard strokes [7 up and 7 down], with 2 min between each pair of strokes to allow heat to dissipate.
The homogenate volume was measured and transferred to a beaker and 1/10 volume of KCl (0.2M) was added dropwise. The solution was stirred on ice for 30 min and then centrifuged at 4°C for 30 min at 17,000 rpm (22,500 x g) in a Sorvall SS-34 rotor. The supernatant was collected and centrifuged at 4°C for 60 min at 37,000 rpm (140,000 x g) in a Beckman 60Ti rotor. The clear middle layer was collected by a pasteur pipette. The top (lipid-rich) and bottom (cellular debris-rich) layers were discarded. The extract was frozen with liquid N2 and stored at -80°C.
The next day the frozen extract was thawed under tap water. The extract was placed in a Spectra/Por 1 dialysis tubing (6000-8000 MWCO) and dialyzed against 1L of buffer D [20mM HEPES (pH 7.8), 20% glycerol, 50mM KCl, 0.2mM EDTA (pH 8), 0.5mM DTT, 0.5mM PMSF] at 4°C, for 3 hrs. The tubing was inverted every 20 min and the buffer was changed after 1 and 2 hrs. The dialyzed mixture was centrifuged at 4°C for 30 min at 17,000 rpm (22,500 x g) in a Sorvall SS-34 rotor. The supernatant was carefully aliquoted into 1.5ml microcentrifuge tubes. The dialyzed extract was frozen in liquid N2 and stored at -80°C
Yeast Prp2 ATPase and its associated G-patch protein
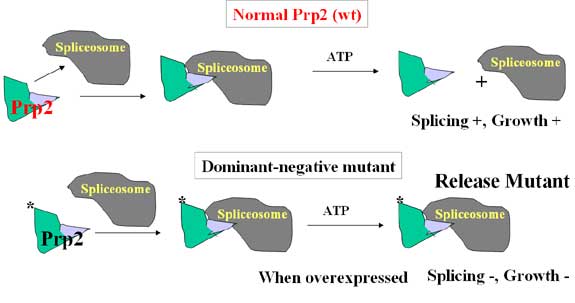
One of the major spliceosomal proteins is RNA helicases, whose function is believed to mediate RNA and protein rearrangement in the spliceosome during the splicing cycle. We characterized one such protein called Prp2 in order to elucidate the function of RNA helicases in splicing. We found that yeast spliceosome could recognize the pre-mRNA in the absence of Prp2 but the complex stalled right before the first step of splicing. The block is released and splicing resumed when Prp2 and ATP were provided, indicating that the spliceosome is not active without Prp2 and the activation process requires ATP hydrolysis catalyzed by a spliceosome-bound Prp2. This activation process is likely conserved through evolution since we recently found that human spliceosome also requires a human protein with great similarity to yeast Prp2 for activity (see project 3).
There are eight RNA helicases in yeast Saccharomyces cerevisiae that are involved in pre-mRNA splicing and each of them is needed for cell viability. These proteins have highly conserved central helicase domain, which is involved in ATP hydrolysis and is absolutely required for its splicing function. The role of the less conserved amino- and carboxyl-extensions, however, remains unclear. We found that the C-terminal domain of Prp2 and a couple of other spliceosomal helicases is required for spliceosome binding. Moreover, a G-patch protein interacting with Prp2 through the C-domain was identified, and we showed that this interaction is essential for the binding of Prp2 to the spliceosome. This helicase-associated protein called Spp2 is required for yeast splicing and plays a role in providing specificity to Prp2 during spliceosome interaction. It remains to be investigated whether additional helicase-associated proteins are involved in the spliceosome binding of other RNA helicases.

Jessica Kurata
Graduate Student
JKurata@coh.org
Ms. Kurata received her B.S. in Joint Biology and Chemistry from Harvey Mudd College. She is currently working toward her Ph.D. at City of Hope’s Irell & Manella Graduate School of Biological Sciences, partially supported by a fellowship from the Frances Berger Foundation. Her research involves using the CRISPR‐Cas9 system to create a genome‐wide library specifically targeting microRNA genes. MicroRNAs are small regulatory molecules important for controlling gene expression, especially during development and tumorigenesis. The CRISPR‐Cas9 library is being used to determine which microRNAs are involved in cancer cell growth or HIV infection. Jessica is also interested in using bioinformatics to extract information from large datasets.

Emilee Bargoma
Graduate Student
EBargoma@coh.org
Ms. Bargoma received her B.S. in Biology with a concentration in Anatomy and Physiology from Cal Poly State University, San Luis Obispo, and an M.S. in Biology from Cal State East Bay, Hayward. Currently working toward her Ph.D. at City of Hope, Emilee focuses on investigating the role of minor (U12‐type) intron splicing factor ZRSR2 in RNA splicing and cell fitness. Using TALEN‐mediated genome editing to generate null alleles of this gene, we found ZRSR2‐knockout cells increased retention of minor introns and decreased cell growth under stress conditions such as serum starvation. Ectopic expression of ZRSR2 or paralogous ZRSR1 restored minor intron splicing as well as cell growth. She will continue to investigate the alterations of the spliceosome and cellular function caused by the loss of ZRSR2. Laboratory techniques and tools such as genome editing, flow cytometry, massive sequencing, and cell analysis are all helping to elucidate the contribution of ZRSR2 in RNA splicing and cell fitness.

LiWang@coh.org
Dr. Wang received her M.D. & Ph.D. in Hematology from Fujian Medical University, Fuzhou, China. Prior to coming to City of Hope, she was a hematologist at the Fujian Medical University Union Hospital, focusing on diagnosis and therapeutics of relapse and refractory leukemia. Her research interests are in disease mechanisms and drug responses in blood malignancy. Currently, she is using deep sequencing and CRISPR/Cas9 gene editing to investigate the role of microRNA in the all‐trans‐retinoic‐acid‐induced differentiation of acute promyelocytic leukemia cells. She also studies the involvement of NPM1 overexpression and mutant nucleophosmin in myelogenous leukemia.
- Skrdlant, L., Stark, J.M., and Lin, R.‐J. (2016). Myelodysplasia‐associated mutations in serine/arginine‐rich splicing factor SRSF2 lead to alternative splicing of CDC25C. BMC Mol Biol. 17:18. PMID: 27552991
- Lin, R.‐J. (2016). RNA‐Protein Complexes and Interactions: Methods and Protocols. In Methods in Molecular Biology series, Humana Press, New Jersey. PMID: 27403468
- Chen, B., Chen, X., Wu, X., Wang, X., Wang, Y., Lin, T. Y., Kurata, J., Wu, J., Vonderfecht, S., Sun, G., Huang, H., Yee, J. K., Hu, J., and Lin, R.‐J. (2015) Disruption of microRNA‐21 by TALEN leads to diminished cell transformation and increased expression of cell‐environment interaction genes. Cancer Lett 356:506‐516. PMID: 25304376
- Zang, S., Lin, T. Y., Chen, X., Gencheva, M., Newo, A. N., Yang, L., Rossi, D., Hu, J., Lin, S. B., Huang, A., and Lin, R.‐J. (2014) GPKOW is essential for pre‐mRNA splicing in vitro and suppresses splicing defect caused by dominant‐negative DHX16 mutation in vivo. Biosci Rep 34:841‐850. PMID: 25296192
- Wang, S.E. and Lin, R.‐J. (2013) MicroRNA and HER2‐overexpressing Cancer. MicroRNA 2:137‐147. PMID: 25070783
- Gencheva, M., Lin, T.‐Y., Wu, X., Yang, L., Richard, C., Jones, M., Lin, S.‐B., and Lin, R.‐J. (2010) Nuclear retention of unspliced pre‐mRNAs by mutant DHX16/hPRP2. J. Biol. Chem., 285:35624‐35632.
- Li, Chunxia, Kato, Mitsuo, Shiue, Lily, Shively, John E., Ares, Manuel, Jr. and Lin, R.‐ J. (2006) Cell Type and Culture Condition‐Dependent Alternative Splicing in Human Breast Cancer Cells Revealed by Splicing‐Sensitive Microarrays. Cancer Res. 66: 1990‐1999.
- Yean, S.‐L., Wuenschell, G., Termini, J., and Lin, R.‐J. (2000) Essential metal ion coordination by U6 small nuclear RNA contributes to catalysis in the spliceosome. Nature, 408: 881‐884. PMID:11130730
- Kim, S.‐H. and Lin, R.‐J. (1996) Spliceosome activation by PRP2 ATPase prior to the first transesterification reaction of pre‐mRNA splicing. Mol. Cell. Biol. 16: 6810‐6819.
- Lin, R.‐J., Lustig, A.J., and Abelson, J. (1987) Splicing of yeast nuclear pre‐mRNA in vitro requires a functional 40S spliceosome and several extrinsic factors. Genes Dev. 1: 7‐18.
- Lustig, A.J., Lin, R.‐J., and Abelson, J. (1986) The yeast RNA gene products are essential for mRNA splicing in vitro. Cell 47: 953‐963.
- Lin, R.‐J., Capage, M., and Hill, C.W. (1984) A repetitive DNA sequence, rhs, responsible for duplications within the Escherichia coli K‐12 chromosome. J. Mol. Biol. 177: 1‐18.