
Nagarajan Vaidehi Lab
Research Lab Overview
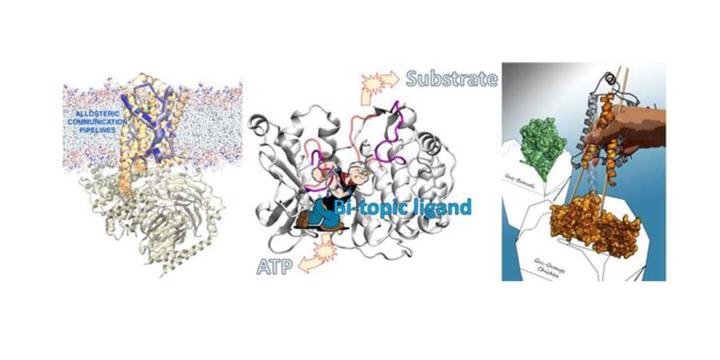
The Vaidehi lab develops physics-based computational methods to study the structure, function and dynamics of proteins and protein complexes. These methods are useful in studying protein-protein interactions and identifying and optimizing small-molecule drugs to inhibit these interactions. Vaidehi’s specific interests are in studying the G-protein-coupled receptor dynamics – as in the interaction of chemokines – and antagonists to the chemokine receptors that trigger leukocyte migration in immune response and inflammation. Prediction of the binding-site interactions of agonists and antagonists with the receptors greatly streamlines the drug development process.
Software
GNEIMO (Generalized Newton-Euler Inverse Mass Operator) is an Internal Coordinate Molecular Dynamics (ICMD) method developed in collaboration with Dr. Abhinandan Jain at the NASA Jet Propulsion Laboratory, Caltech.
The GneimoSim software can be downloaded free of cost for academic use by submitting the request form shown below. Once the form is submitted we will send you a downloadable link to the software.
Allosteer: This software used molecular dynamics trajectories and calculates the network of amino acid residues involved in allosteric communication between two specified sites in a protein. It also calculates the pipeline of this communication and lists all the residues involved in this communication pipeline. To download Allosteer for non-commercial academic use please submit the request form below. Once the form is submitted we will send you a downloadable link to the software.
The Chair of the Department of Computational and Quantitative Medicine, Nagarajan Vaidehi conducts research that focuses on developing physics-based computational methods to study protein structure, dynamics and drug design.



- B.A., Chemistry, Hanyang University, Seoul, Korea
- M.S., Physical Chemistry, Hanyang University, Seoul, Korea
- Ph.D., Physical Chemistry, University of Florida
Research interests:
- Development of biased ligand on angiotensin II type 1 (AT1R) GPCRs
- Design of novel milder detergent for thermostabilized GPCRs
Email: salee@coh.org


Education:
- B.Sc., Chemistry, St. Xavier's College (autonomous) Kolkata, West Bengal, India
- M.Sc., Applied Chemistry, Indian Institute of Technology Dhanbad, Jharkhand, India
- Ph.D., Physical Chemistry, Indian Institute of Technology Bombay, Maharashtra, India
Research interests:
- Identifying thermostable point mutations for GPCRs using energy scores and evolutionary constraints, aggregation propensity of chemokine receptors using classical MD simulations.
Email: sghosh@coh.org

Education:
- B.E., Chemical Engineering, Jadavpur University, Kolkata, India
- M.Tech., Chemical Engineering, Indian Institute of Technology Guwahati, Guwahati, India
- Ph.D., Chemical Engineering, University of Kentucky, KY, USA.
Research interests:
- To develop and modify computational chemistry tools that can rapidly predict thermostabilizing mutations for membrane receptors, which is a critical step for receptor crystallization
- Understanding the stability of membrane receptors in various detergent micelles, important for protein extraction and purification.


Education:
- B.S., Biology, Henan Normal University, Xinxiang, China
- M.S., Microbiology, Huaqiao University, Quanzhou, China
- Ph.D., Computational Chemistry, University of South Florida, Florida
Research interests:
- Kinase dynamics simulation, GPCR-membrane simulation, conformational free energy simulation, method development.
Email: nma@coh.org


Education:
- B.Tech., Bioinformatics, Vellore Institute of Technology University, Tamil Nadu, India
- Ph.D., Computational Chemistry, University of Georgia
Research interests:
- Biased agonism using classical MD simulations, biased ligand design, studying the effects of allosteric communication in GPCRs using MD and AlloSteer, understanding the molecular basis for subtype selectivity and method development.
Email: anivedha@coh.org


Education:
- B.Sc., Bioengineering, University of California, Riverside
Research interests:
- Applying machine learning in the study of GPCR thermostability
Email: smuk@coh.org


Education:
- B.Sc., Biomedical Engineering at Eindhoven University of Technology (TU/e)
- M.Sc., Biomedical Engineering at Eindhoven University of Technology (TU/e)
Research interests:
- Investigating GPCR c-term binding to G-protein through molecular dynamics simulations
Email: phendrikx@coh.org


Education:
- B.A., Biology, University of California, Santa Barbara
- M.S., Biochemistry, California State University, Northridge
- Ph.D. (in progress), Biology, Irell & Manella Graduate School of Biological Sciences, City of Hope, Duarte, California
Research interests:
- Studying the molecular mechanisms of partial agonism and allostery in G protein coupled receptors, and how each process influences G protein selectivity3
Email: spaulekas@coh.org


Education:
- B.S., Human Biology, University of California, San Diego
- Ph.D., (in progress), Computational Biophysics, Irell & Manella Graduate School of Biological Sciences, City of Hope, Duarte, California
Research interests:
- Delineating the structural features of the GPCR-G protein complex that lead to selective signaling to different G protein pathways
Email: msandhu@coh.org
We collaborate with organizations in progressing the development of new treatments in our specialized areas of research.

Nivedha, A.K., Tautermann, C.S., Bhattacharya, S., Lee, S., Casarosa, P., Kollak, I., Kiechle, T. and Vaidehi, N.
Li, H., Sandhu, M., Malkas, L.H., Hickey, R.J. and Vaidehi, N.
Lee, S., Devamani, T., Song, H.D., Sandhu, M., Larsen, A., Sommese, R., Jain, A., Vaidehi, N. and Sivaramakrishnan, S.
Latest Research News
34.1293487, -117.9726643
Duarte, CA 91010


