Areas of Research
The Division of Mathematical Oncology and Computational Systems Biology is engaged in numerous areas of collaborative research. See the following recent publications for summaries of the field:
- The 2019 Mathematical Oncology Roadmap (Physical Biology)
- Introduction to Mathematical Oncology (JCO CCI)
Areas of active research at City of Hope include:
- Modeling tumor/immune system dynamics; specifically how cancer cells evolve to hide from immune cell recognition, and if that process can be halted or reversed
- Applying evolutionary theory to understand leukemia development, particularly the growth and proliferation of treatment-resistant “leukemia stem cells”
- Using imaging data, including positron emission tomography, or PET, and magnetic resonance imaging, or MRI, data to help predict a cancer’s growth and response to treatment, particularly for cancers such as brain tumors that are hard to observe directly
- Integrating complex data sets to optimize therapies for best clinical and quality-of-life outcomes
- Improve screening and early detection efforts by identifying and quantifying factors that contribute to cancer risk
- Enhancing clinical trials through better selection and enrollment of candidates — patients who are most likely to benefit from the tested therapy
Investigating the biomechanical impact of tumor growth
Brain tumors present with different growth phenotypes, ranging from invasive lesions without notable ‘mass effect’ to strongly displacing lesions that induce mechanical stresses and result in healthy tissue deformation, midline shift or herniation. Biomechanical forces, such as those resulting from displacive tumor growth, shape the tumor environment and are known to affect tumor growth and evolution. Likewise, tumor growth drives physical changes in the micro-environment that affect tissue solid and fluid mechanics.

We develop mathematical models to study the biomechanical impact of brain tumor growth and possible treatment implications. We use these models to investigate determinants of tumor shape, such as the influence of tissue structure and growth location, and to evaluate quantitative image-based measures of tumor mass effect. Along with these models, we develop approaches for identifying patient-specific growth characteristics from clinical imaging data.
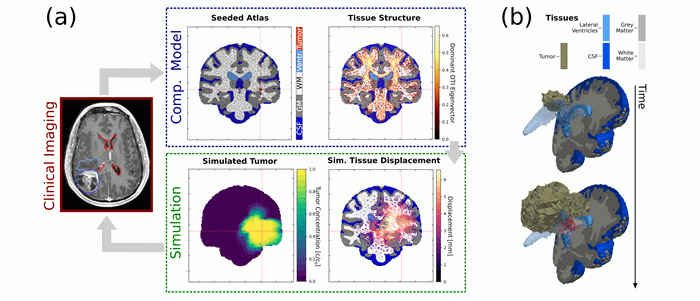
Simulation of brain tumor growth and mass effect: (a) Computational models are initialized from clinical imaging data. The simulation predicts tumor invasion and mechanical stresses induced by tumor growth. Simulation results are compared to imaging findings. (b) Simulated time evolution of 3D tumor growth and tissue deformation. Supported by the European Commission Horizon 2020 ‘GlimS’ grant.
Modeling migration of cell-based therapies for high grade glioma
Neural stem cells (NSCs) can potentially be used for therapy in scenarios of brain tumors and traumatic brain injuries where they can act as drug delivery vehicles or demonstrate their cell regeneration and tissue repair capabilities. The route of cell migration from an injection site to pathological site can affect the migration and thus therapeutic efficiency of these cells. We use structure tensor analysis of tissue white matter to model the migration of neural stem cells across the brain. The application of the model on two-dimensional histological sections is shown below.
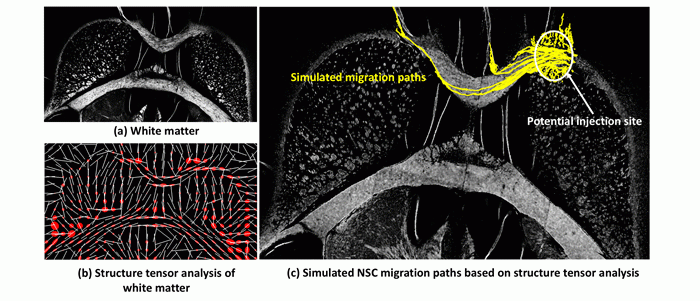
Simulated migration paths of NSCs. NSC migration paths are simulated from potential injection site in the corpus callosum. The image shows (a) an image of tissue white matter imaged on histological section. (b) Structure tensor analysis of the white matter image showing the dominant eigenvectors (white lines) and tensor ellipses (red). (c) Simulated paths in yellow overlaid on top of an image of tissue white matter by modeling cell migration based on dominant eigenvector directions. 300 cells were initialized in silico within the potential injection site region. Cells are seen to move along the white matter where the eigenvectors are aligned with each other at different points. Cells in the gray matter remain close to the injection site. The application of this model on immune-histological images and analysis of migration patterns of these cells can be seen here.
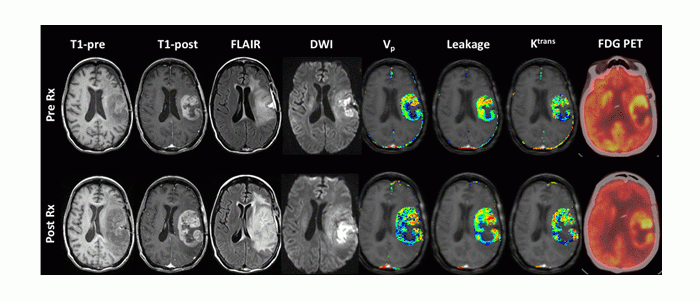
Multimodal (MRI, PET), multiparametric (perfusion, diffusion) imaging of response to cell-based therapy in high grade glioma. Early Changes in Tumor Perfusion from T1-Weighted Dynamic Contrast-Enhanced MRI following Neural Stem Cell-Mediated Therapy of Recurrent High-Grade Glioma Correlate with Overall Survival
Mathematics and mathematical structures of biological space-time
We are interested in theoretical and mathematical perspectives on the process of aging. We extend the concepts of physical space and time to an abstract, mathematically-defined space, which we associate with a concept of “biological space-time” in which biological dynamics may be represented. We hypothesize that biological dynamics, represented as trajectories in biological space-time, may be used to model and study different rates of biological aging. As a consequence of this hypothesis, we show how dilation or contraction of time analogous to relativistic corrections of physical time resulting from accelerated or decelerated biological dynamics may be used to study precipitous or protracted aging with an emphasis on cancer in aging.
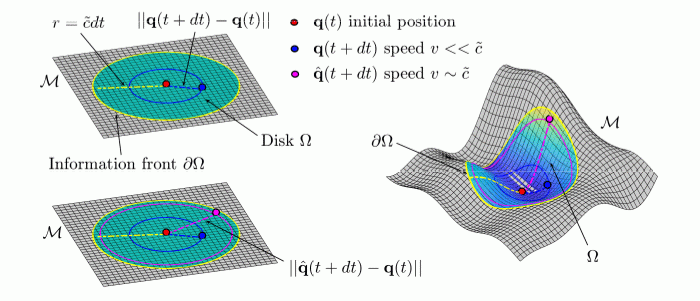
Representation of an information front propagating from the center of a disk on a flat plane M in a biological space-time. Information can travel no faster than c ̃. The distance traveled by a point in the interval of time dt is given by ||q(t + dt) − q(t)|| and it will be always less than the maximum speed of information traveled over the same time interval. For biological dynamics occurring at near the speed of the information limit, biological time may dilate. Aging in a Relativistic Biological Space-Time
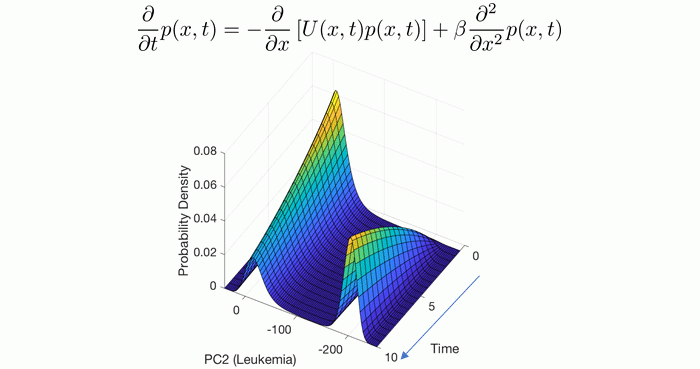
Modeling cancer as a state transition of the transcriptome
Viewing gene expression dynamics as a stochastic process allows us to interpret the spatial-temporal evolution of the probability density function with the Fokker-Planck equation. Shown is the solution of this partial differential equation with parameters estimated from data which predicts the time to develop leukemia from a given initial configuration of gene expression. State-Transition Analysis of Time-Sequential Gene Expression Identifies Critical Points That Predict Leukemia Development
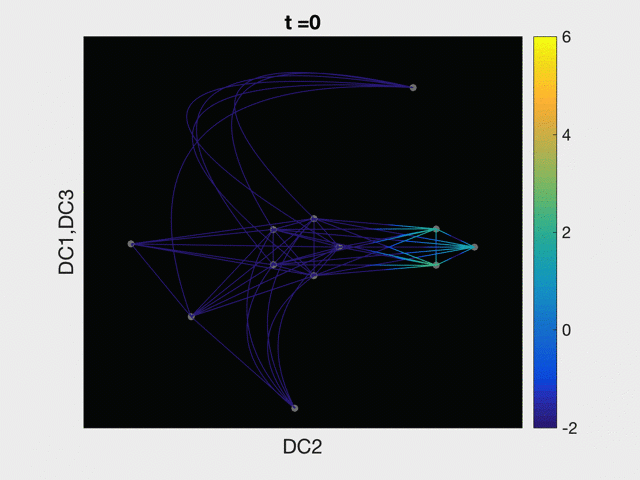
Modeling with single-cell RNA-sequencing data
The advent and pervasive use of single cell sequencing technologies has brought our study of disease to the single cell level. Here we model the process of both normal and abnormal cellular differentiation with partial differential equations solved on a graph abstracted from single cell RNA-sequencing data. Modeling Acute Myeloid Leukemia in a continuum of differentiation states
For more information about Mathematical Oncology research, or if you are interested in partnering with the division for a study, please contact: